Override a single method to plug in your own network architecture. Grab a tool from the box and customize your experiments.
- Sliding window
- ROI inspection & cropping
- Automatic padding & shape alignment

MIP Candy brings ready-to-use training, inference, and evaluation pipelines together with aesthetics, so you can focus on your experiments, not boilerplate.




MIP Candy powers research across top institutions.
Designed for modern medical image research pipelines.
Override a single method to plug in your own network architecture. Grab a tool from the box and customize your experiments.
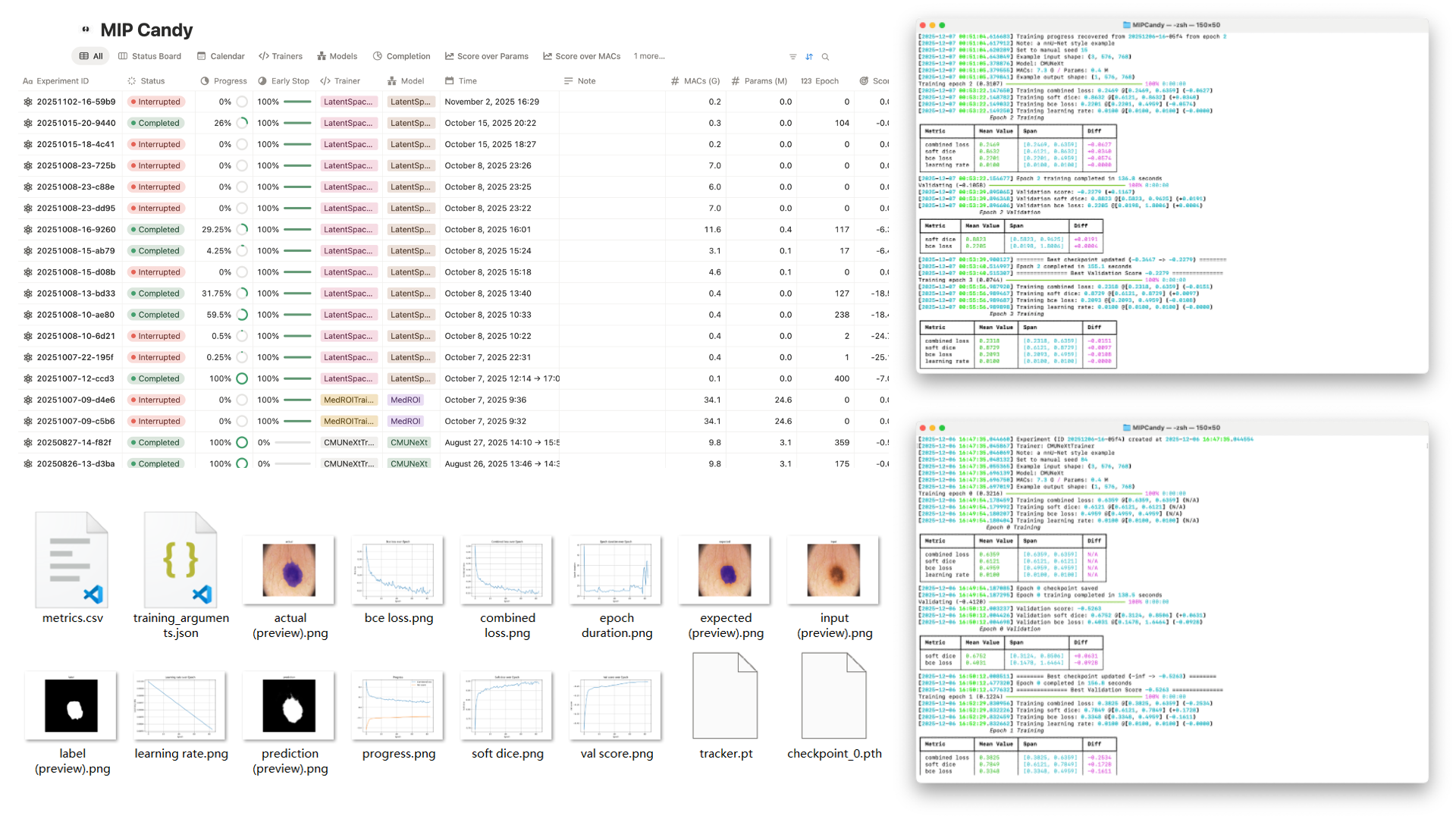
A clean CLI layout makes it easy to configure experiments, track progress, and resume work without digging through scripts.
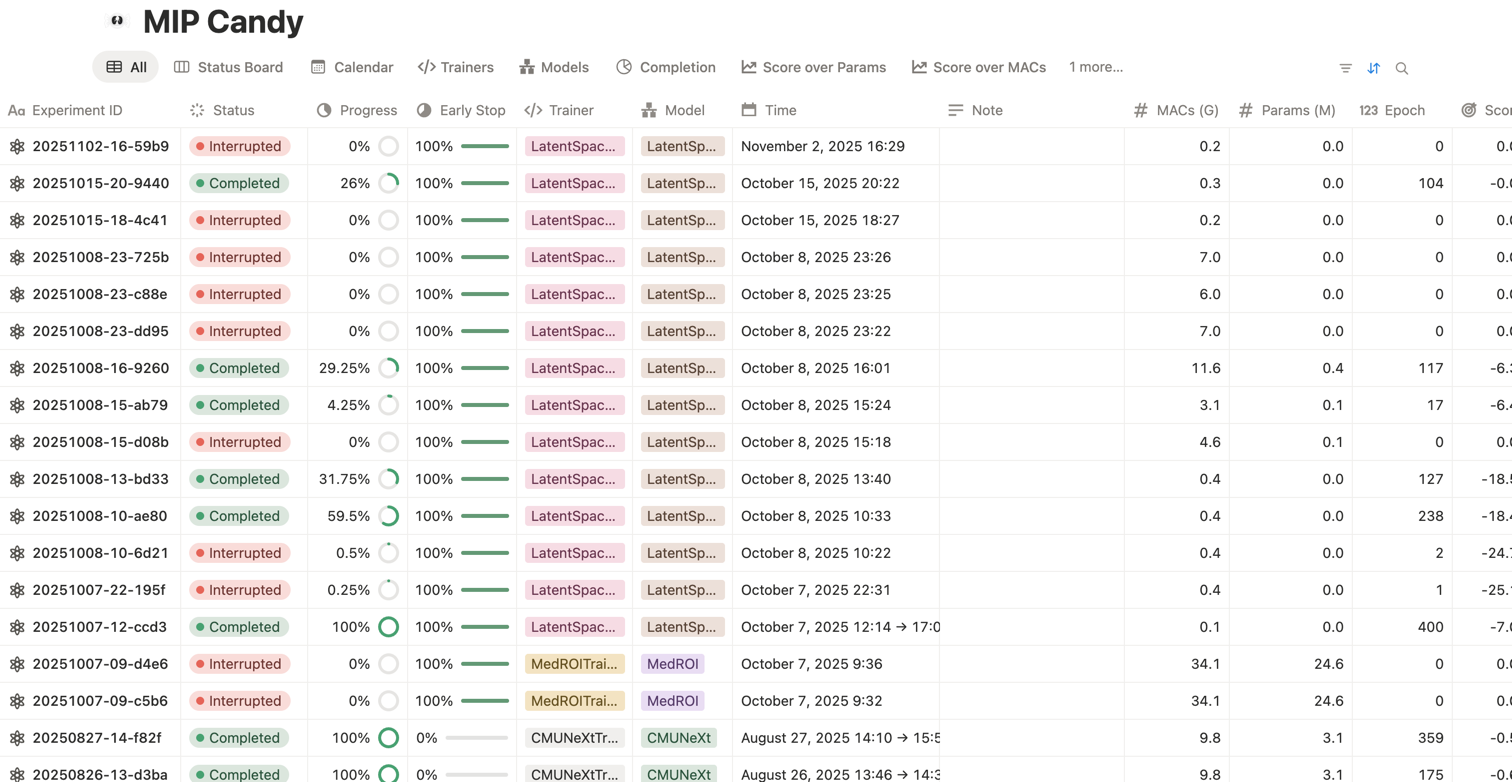
Inspect slices or volumes directly from the training pipeline for intuitive understanding of your data and predictions.

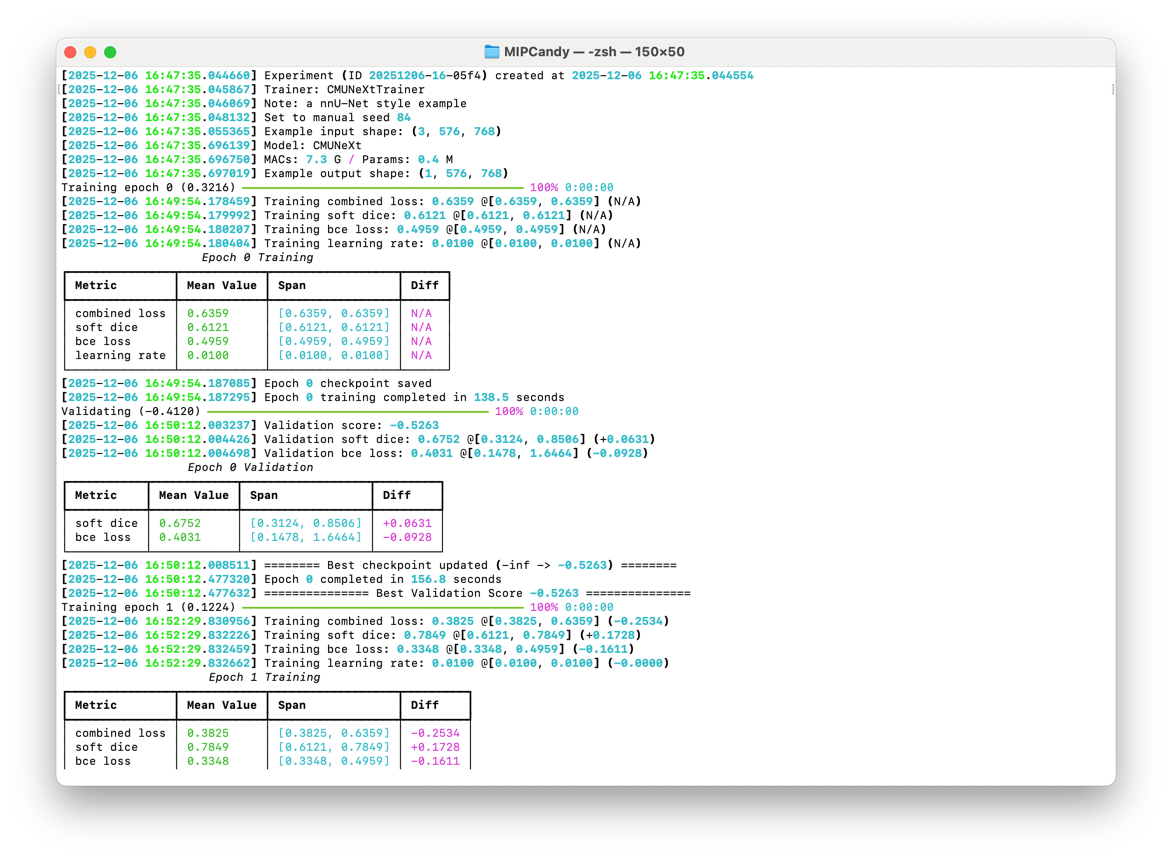
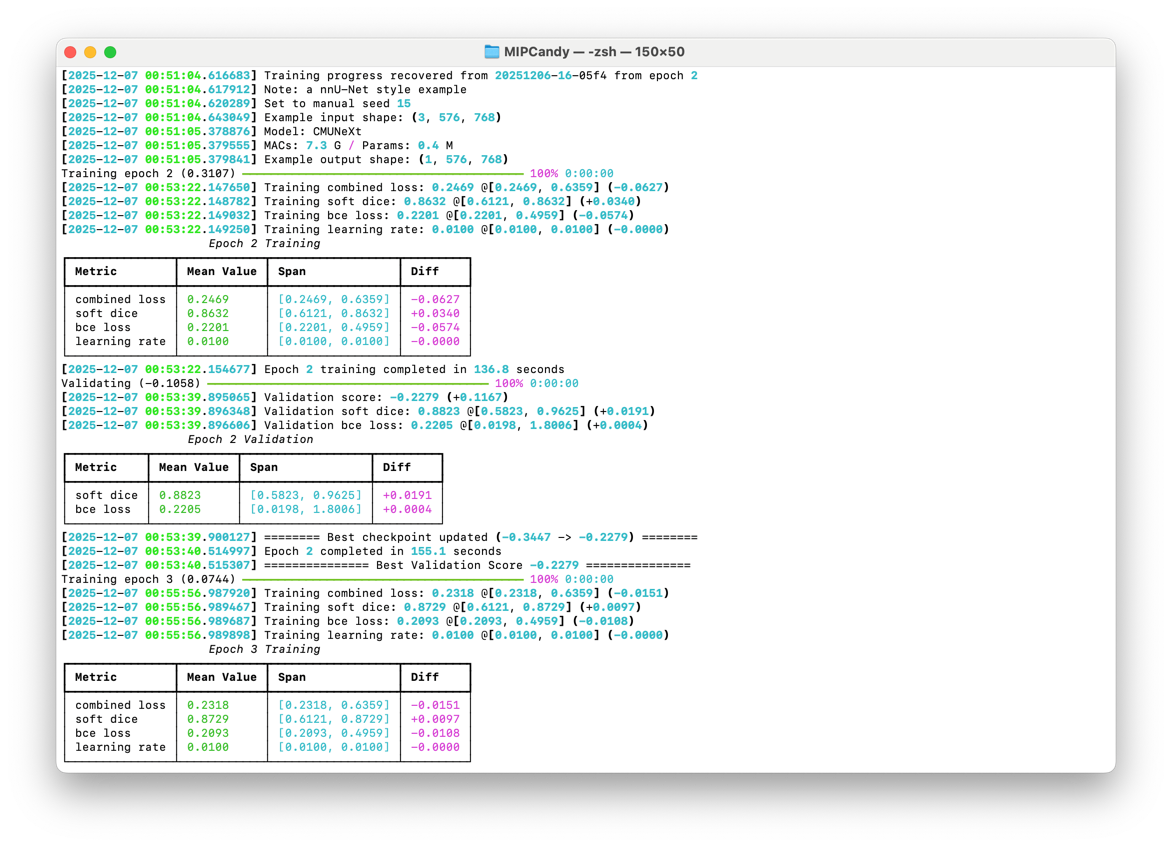
Experiments can be safely paused and resumed with built-in recovery mechanisms, so cluster hiccups don’t cost you progress.
Connect to Notion, Weights & Biases, and TensorBoard for rich experiment tracking and sharing with your team.
Built for Python 3.12 and above, MIP Candy takes advantage of modern typing and ecosystem improvements out of the box.
Start from SegmentationTrainer and only implement the network construction. MIP Candy handles data flow, loss computation, augmentation, checkpointing, and evaluation out of the box.
from typing import override
from torch import nn
from mipcandy import SegmentationTrainer
class MyTrainer(SegmentationTrainer):
@override
def build_network(self, example_shape: tuple[int, ...]) -> nn.Module:
...
Explore an interactive MIP Candy frontend demo directly in Notion.
Download a dataset, create a dataset wrapper, and hand it to a trainer. Below is an example using the PH2 dataset. To replicate an nnU-Net style training, you can add data augmentations and use the provided SlidingSegmentationTrainer instead of the current SegmentationTrainer to utilize the sliding window mechanism.
from typing import override
from monai.networks.nets import BasicUNet
from monai.transforms import Resized
from torch import nn
from torch.utils.data import DataLoader
from torchvision.transforms import Compose
from mipcandy import SegmentationTrainer, AmbiguousShape, download_dataset, JointTransform, MONAITransform, Normalize, \
NNUNetDataset
class UNetTrainer(SegmentationTrainer):
@override
def build_network(self, example_shape: AmbiguousShape) -> nn.Module:
return BasicUNet(2, example_shape[0], self.num_classes)
download_dataset("nnunet_datasets/PH2", "tutorial/datasets/PH2")
transform = JointTransform(transform=Compose([
Resized(("image", "label"), (560, 768)), MONAITransform(Normalize(domain=(0, 1), strict=True))
]))
dataset, val_dataset = NNUNetDataset("tutorial/datasets/PH2", transform=transform, device="cuda").fold()
dataloader = DataLoader(dataset, 8, shuffle=True)
val_dataloader = DataLoader(val_dataset, 1, shuffle=False)
trainer = UNetTrainer("tutorial", dataloader, val_dataloader, device="cuda")
trainer.train(1000, note="example with PH2 dataset")
MIP Candy requires Python ≥ 3.12. Install the standard bundle from PyPI:
MIP Candy Bundles provide verified model architectures with corresponding trainers and predictors. You can install it along with MIP Candy.
Jump into the ecosystem around MIP Candy.